
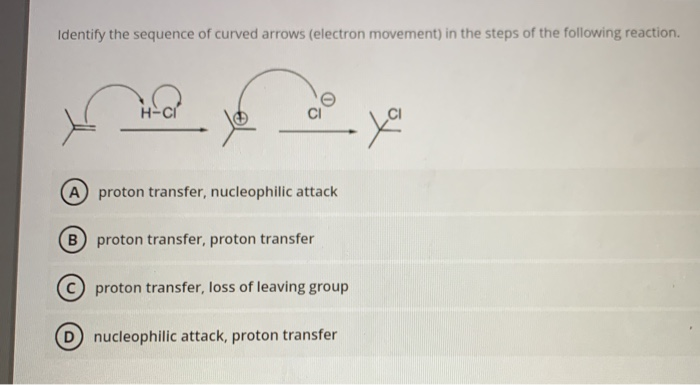
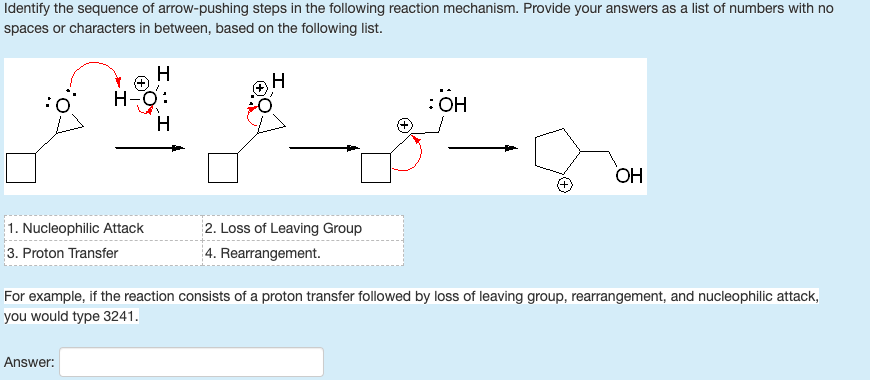
But it is easier to use this Rule: x n n (n+1)/2. 3 4 5 Sequence boundaries are associated with intervals of few or no in situ fossils and often abundance peaks of reworked fossils in the. In addition to CPuORFs, the pipeline was able to find 14 conserved coding regions directly upstream and in frame with the main ORF, which likely initiate translation on a non-AUG start codon. The Triangular Number Sequence is generated from a pattern of dots which form a triangle: By adding another row of dots and counting all the dots we can find the next number of the sequence. Identifying whether an oil palm plantlet is the desired shell type can take six years, and, by that time, the trees cannot be uprooted. In the Texas offshore Pliocene and Pleistocene depocenter, patterns of fossil abundance are often the most widely applicable observational criteria for identifying the surfaces that define sequences. Our new pipeline was able to identify 30 novel Arabidopsis CPuORFs, conserved across a wide variety of eudicot species of which 16 do not initiate with an AUG start codon. For this reason, we developed a novel pipeline to identify CPuORFs unbiased of start codon using well annotated sequence data from 32 eudicot plant species and rice. Finding Missing Numbers To find a missing number, first find a Rule behind the Sequence.
#IDENTIFIES SEQUENCES SERIES#
Each number in the sequence is called a term (or sometimes 'element' or 'member'), read Sequences and Series for a more in-depth discussion.
Sequences with amino acid repeats are referred to as low complexity. Sequence A Sequence is a set of things (usually numbers) that are in order. The suboptimal initiation context may even include non-AUG start codons, which makes CPuORFs hard to predict. The study, published in Nature Communications, identifies the exciting role of the. Previous research has shown that most CPuORFs possess a start codon context suboptimal for translation initiation, which turns out to be favorable for translational regulation. To this end, comparative genomic approaches have been used to identify novel CPuORFs, by comparing AUG initiating uORF sequences of the Arabidopsis genome or Arabidopsis ESTs. read more to regulate the translation of the downstream main ORF, usually in a metabolite concentration dependent manner. Most experimentally validated CPuORFs demonstrated. If the duplicate rule identifies the record as a duplicate and uses the block action, the record isnt saved and no further steps. An unknown fraction of these uORFs encode sequence conserved peptides (conserved peptide uORFs, CPuORFs). Eukaryotic mRNAs contain a 5' leader preceding the main open reading frame (mORF) and, depending on the species, 20-50% of eukaryotic mRNAs harbor an upstream ORF (uORF) in the 5' leader.


 0 kommentar(er)
0 kommentar(er)
